Now almost a year into a two-year project, Dr Bibek Gooptu, from the Institute for Structural and Molecular Biology (ISMB) at Birkbeck and UCL, and his team are researching new treatments for the condition Alpha-1 Antitrypsin Deficiency and helping to advance our understanding of the mechanisms behind the condition.
The protein Alpha-1 Antitrypsin is created in the liver and then carried in the blood to the lungs, where it protects lung tissue. In people with Alpha-1 Antitrypsin Deficiency, a mutant gene within the liver cells causes the Antitrypsin protein to adopt a different structure, and individual protein molecules link together in chains, known as polymers. The liver is not able to secrete these polymers and so they remain stuck inside liver cells, damaging them. It also means that not enough Antitrypsin reaches the lungs from the bloodstream. People with Antitrypsin Deficiency are likely to suffer both from liver disease (e.g. cirrhosis or cancer of the liver) and, if they are smokers, they are likely to develop emphysema at a much younger age than someone without Antitrypsin Deficiency – probably in their 30s or 40s.
Promising compounds
Dr Gooptu and the project team (Gooptu group and collaborators at Birkbeck, UCL and Cambridge) have been studying compounds which will bind to the Antitrypsin protein in such a way that it is prevented from forming polymers. Because they have previously solved the molecular structure of the Antitrypsin in atomic detail they have identified three possible drug binding sites to target with new treatments. They have confirmed that small molecules (drug building blocks) predicted to target these sites in computer simulations bind the protein when tested experimentally in solution. The team are currently finalising computer simulations identifying a set of 750 compounds with similar binding characteristics to these ‘hits’. They will use these to test run the technology they are developing to assess the effects of many molecules at a time, not just on the protein in a test tube (which they do using a technique called mass spectrometry) but also at the site where the disease starts: in cells.
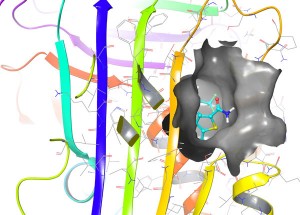
Example of a compound (cyan) successfully binding one of the target sites (surface features shown in grey) in Alpha-1 Antitrypsin (molecular structure shown as coloured ribbons) in computer simulation.
Testing in mammalian cells
Although a compound may bind to a protein in a solution, to be a successful drug the compound has to be able to enter the cell effectively. Once inside it must bind effectively in conditions that may be quite different. To see whether the compounds which bound successfully in solution can bind in the same way in cells like liver cells the team are testing them in cells known as CHO cells. These are simple mammalian cells that grow well in a nutrient broth (cell culture) and behave like liver cells in terms of how they deal with normal Antitrypsin (secreting it efficiently) and disease mutant forms (accumulating polymers within the cell). It is already possible to check these cells for beneficial effects of drug-like compounds by checking whether the amount of secreted Antitrypsin goes up and/or the polymer levels go down. However at early stages of drug development the effects of the compounds may be more subtle, so Dr Gooptu and his post-doctoral researcher, Dr Nyon, are working with the Thalassinos group at UCL to identify more sensitive markers. For this they are working out the molecular signatures of health and disease in the part of the cell where proteins are prepared for secretion and where Antitrypsin polymers form and get stuck. These signatures are the levels of different proteins found within this area, known as the endoplasmic reticulum or ER for short. Not only is this useful for the current project, the signatures may themselves provide clues for other treatment options in the future.
Other proteins can aggravate or lessen disease
As well as the Antitrypsin protein there are hundreds of other proteins present in the ER. The amounts in which these proteins are present change depending on whether the cell is healthy or unhealthy. Some drop to undetectable levels while some new proteins are only found in one or other situation. Dr Gooptu compares this to looking at a street from an aerial view. In one view there are people in the street, with music and bunting. This is a street party and the equivalent to the ER of a healthy cell. In another view of the same street there might again be people in the street, but this time you also observe flames, fire engines and hoses. This is an emergency situation and the equivalent of the ER in an unhealthy cell. However, it is necessary to establish whether the people (proteins) in the second scene are causing the disease (arsonists) or fighting it (fire fighters). This will allow comparison of desirable with harmful responses to promising compounds and identify other proteins that might themselves be useful targets for future drug treatments.
Understanding how compounds are binding
Once they have identified the best compounds from the original set of 750 that bind Antitrypsin both in free solution and within cells, a technique known as Nuclear Magnetic Resonance (NMR) spectroscopy will be used to look very closely at the binding process and discover exactly how it takes place.
Binding of the Antitrypsin protein (grey ribbon representation) to a prototype drug molecule can be followed by NMR looking at changes seen when the interaction occurs around the structure at many individual points (spheres). In this case the colour coding shows many areas that change a lot (blue), whilst a few areas are stable (white)
Using this technique the team can identify the characteristics of the protein:compound interaction. Armed with this information they can return to a library of thousands of compounds and identify further potential binders for testing and develop compounds which will bind in the most effective way.
Improving understanding of Alpha-1 Antitrypsin deficiency and the disease mechanisms
The approach taken by Dr Gooptu has developed existing methods and combined them in a new way. Usually drug companies will blind-test thousands of compounds in solution. Using the computational modelling before the in-solution testing meant that the team could identify a smaller number of compounds which were more likely to give positive results. The more detailed NMR spectroscopy studies are then targeted on a small number of compounds that show the most promise both in solution and in cells. This approach also has the additional advantage that while identifying compounds that bind Antitrypsin it also reveals more about how the genetic mutation causes disease in terms of harmful changes in proteins and cell responses.
Next steps
Having carefully developed the individual computational, mass spectrometry, cell biology and NMR techniques over the last year, in the next 12 months the project team will put them together and see how well they work in a pipeline for assessing the 750 test compounds. If this works well the pipeline could then be boosted to screen far greater numbers of compounds in future. However above the molecular scale, the project has already paved the way for bigger drug discovery projects in Alpha-1 Antitrypsin deficiency. Through his work, and the forum of research meetings convened by the US patient charity that funds the project (the Alpha-1 Foundation), Dr Gooptu has established contact with other groups working on a range of other approaches to identify new treatments. He is now collaborating with these groups and the Foundation to develop larger screens in which hit molecules can be rapidly identified from cell screening. The hits will then be studied in parallel by his group in Birkbeck and US research groups with complementary expertise to establish how they work and so how they can be further improved as efficiently as possible.
Dr Bibek Gooptu has been working on Antitrypsin Deficiency at Birkbeck since 2006. He is also a practising Consultant in Respiratory Medicine. His research is supported by the Alpha-1 Foundation, the Medical Research Council and the Wellcome Trust.


Great to read this. Thank you for carrying out this research. As someone newly diagnosed as PiZZ with a level of 0.21 it is good to know that there is some hope for treatment in the future. I have also shared this with my sister, also PiZZ 0.34.
Msg for those Alpha’s who don’t already know – there is an online support group who have a website with loads of info here:
http://www.alpha1.org.uk/